The safety and efficiency of CRISPR-based genome editing may be improved by using an enzyme that cuts just one strand of DNA instead of both.
A research team at the University of California, San Diego (UCSD), found that swapping the bacterial enzyme that is traditionally used with CRISPR, Cas9, for another, called nickase, significantly reduced off-target effects.
Lead author Dr Sitara Roy, said 'I could not believe how well the nickase worked – it was completely unanticipated.'
'In general, with existing CRISPR techniques, you have to worry about roughly one percent of edits being mistakes or off-target,' added co-senior author, Professor Ethan Bier. 'I would say that, in the case of our system, it would be more like one in 10,000'.
The CRISPR approach to genome editing has revolutionised modern research. It usually relies on Cas9 to cut through both strands of DNA, but this is thought to have just 20 to 30 percent efficiency, and often causes off-target effects.
The team were investigating using CRISPR in combination with a natural DNA repair mechanism called homologous chromosome-templated repair (HTR) to correct mutations.
'The healthy variant can be used by the cell's repair machinery to correct the defective mutation after cutting the mutant DNA,' explained senior co-author Dr Annabel Guichard.
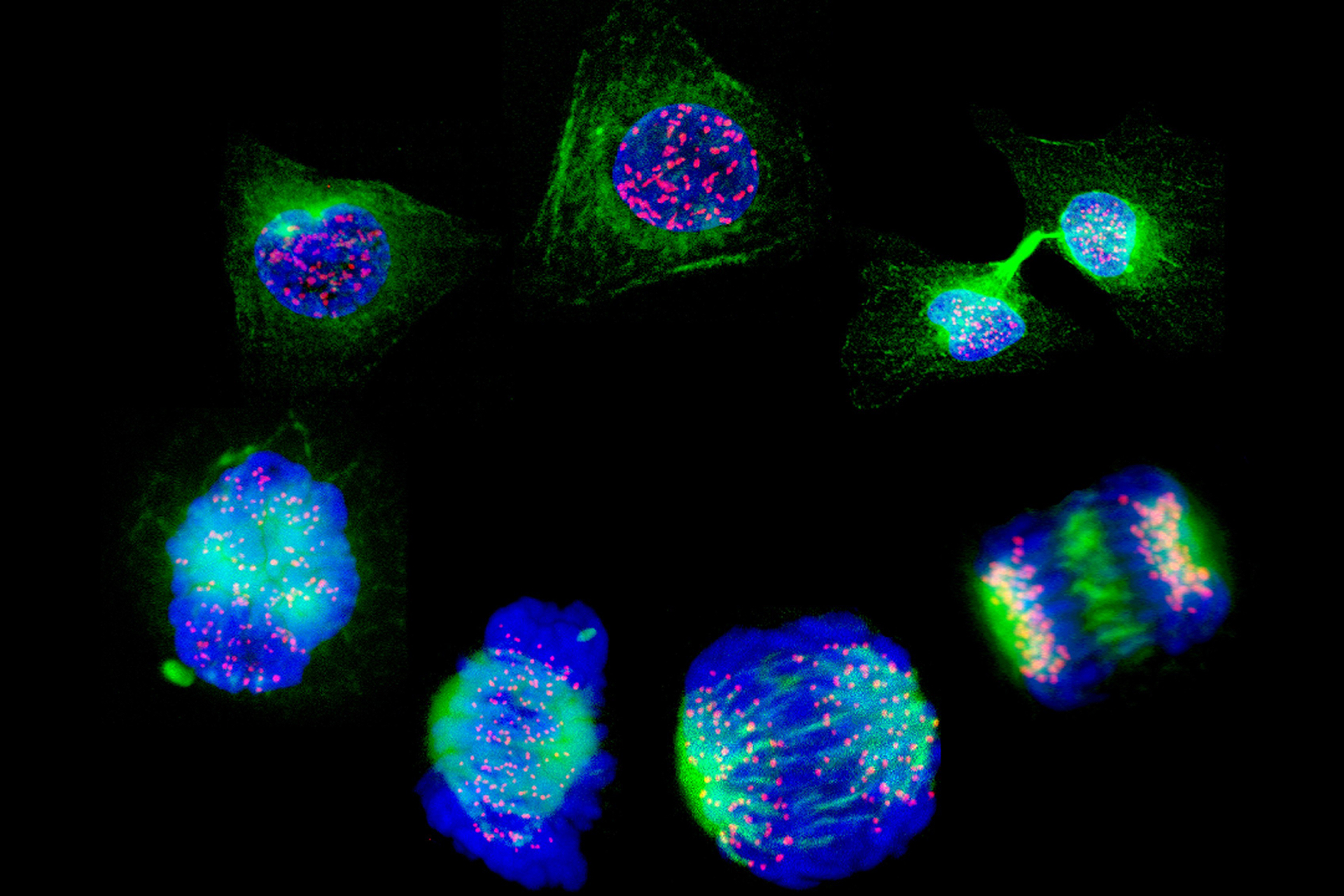
The team at UCSD used Drosophila fruit flies, to test the best types of DNA breaks for HTR to work. They evaluated the safety and efficiency of repairing double-strand DNA breaks – the breaks that Cas9 induces – and single-strand DNA breaks, which nickase relies on.
HTR was able to complete the repair in 40 to 65 percent of cuts made using the Nickase enzyme, compared to the 20 to 30 percent of cuts made using the Cas9.
Although this work was performed in flies, the team are hopeful it will also work in mammals.
'We don't know yet how this process will translate to human cells and if we can apply it to any gene,' added Dr Guichard. 'Some adjustment may be needed to obtain efficient HTR for disease-causing mutations carried by human chromosomes.'
The study was published in Science Advances.
Sources and References
-
Cas9/Nickase-induced allelic conversion by homologous chromosome-templated repair in Drosophila somatic cells
-
'Soft' CRISPR may offer a new fix for genetic defects
-
'Softer' form of CRISPR may edit genes more accurately
-
New 'soft' CRISPR nicking system simple, efficient, and safe
-
New CRISPR technique causes few unintended mutations in fruit flies



Leave a Reply
You must be logged in to post a comment.