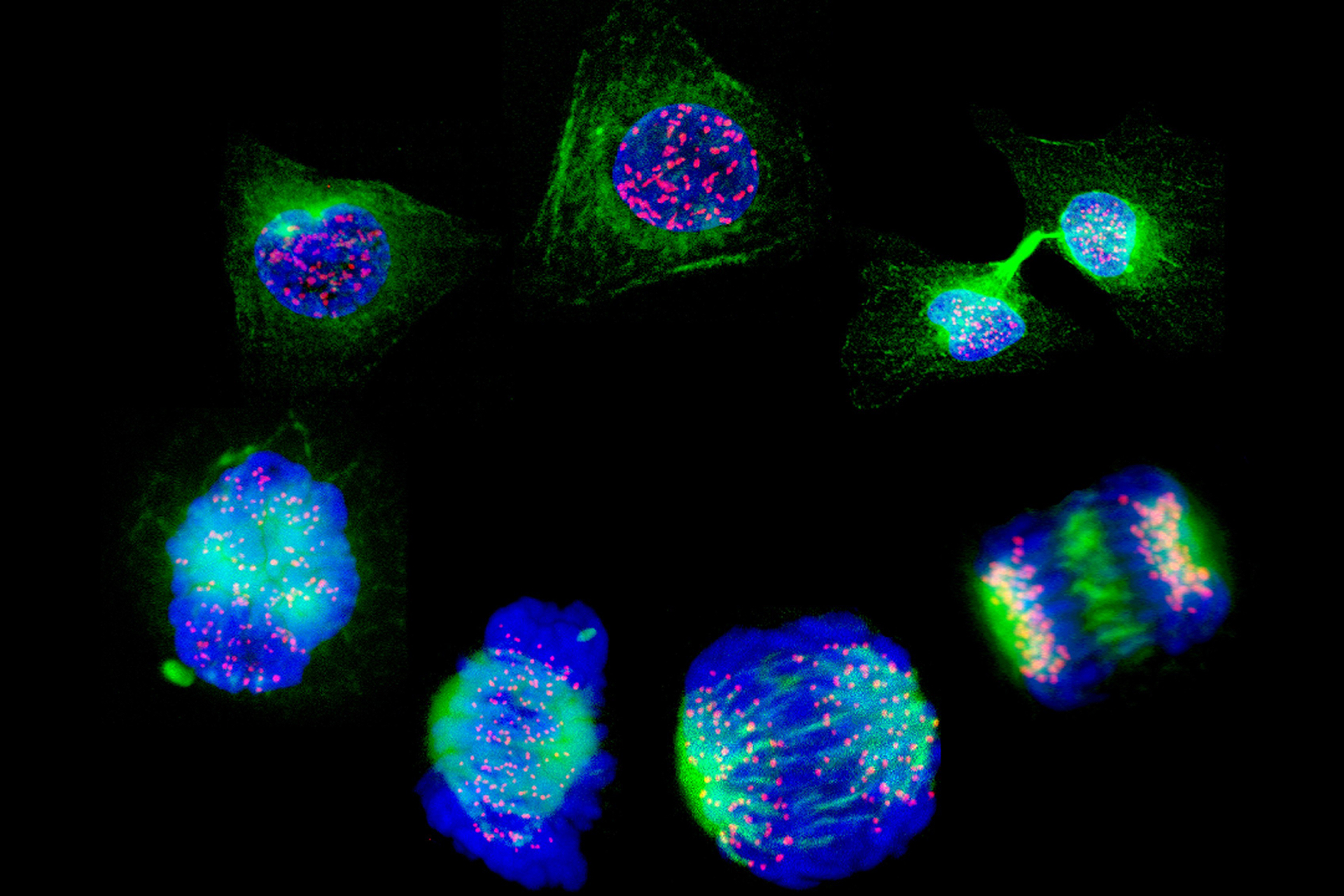
A genome editing technique that allows researchers to test thousands of different mutations that might affect how cancer cells behave has been developed.
Massachusetts Institute of Technology (MIT) researchers used this method to screen mutations in the tumour-suppressor gene TP53, which is frequently mutated in cancer. Unlike earlier technologies however, this new technique allows researchers to insert the mutations found in patient tumours into the cells' own copies of the TP53 gene, rather than introduce new modified genes. This allowed them to discover known mutations with functional consequences, which previous methods had missed.
Dr Francisco Sanchez-Rivera, assistant professor of biology at MIT, and senior author of the study said: 'In one experiment, you can generate thousands of genotypes that are seen in cancer patients, and immediately test whether one or more of those genotypes are sensitive or resistant to any type of therapy that you're interested in using'.
Reported in Nature Biotechnology, the study uses a modified CRISPR approach called 'prime editing' which can be used to make almost any desired small-scale DNA change. Unlike using CRISPR/Cas, where a gene is deactivated by cutting both DNA strands, prime editing uses engineered enzymes to carry a short strand of RNA to a specific target region in the genome, where it 'over-writes' the corresponding DNA sequence.
The key technical innovation in this study overcomes a major limitation which previously prevented large scale prime editing screens; the different RNA sequences (or 'guides') used to target genes are extremely variable in their ability to successfully cause their intended DNA change, despite recent improvements such as using machine learning predictions (see BioNews 1180). The authors surmounted this by simultaneously introducing DNA sequences which match each target site. These 'sensor' sequences can be easily checked for editing, permitting the researchers to focus on the signal and filter out the noise.
Traditional comparable experiments involve adding a modified version of a gene with the desired mutation to cells, which usually results in producing far greater amounts of protein than normal ('over-express'). However, such techniques failed to identify several TP53 mutations highlighted in this approach which appear to affect the ability of the protein to form its natural complexes, as the over-abundance of protein could counteract what the proteins would be doing at normal levels.
'There are many variants in p53 [the protein encoded by TP53] that remain understudied' said Samuel Gould, the MIT graduate student who lead this work, '… this is a case where you could only observe these variant-induced phenotypes if you're engineering the variants in their natural context and not with these more artificial systems'.
While this study focuses on one gene, the authors report how it can be deployed to make a variety of different changes to a cancer cell's genome to answer various questions.
Sources and References
-
Scientists develop a rapid gene-editing screen to find effects of cancer mutations
-
High-throughput evaluation of genetic variants with prime editing sensor libraries
-
Prime editing approach provides a faster, more effective way to screen for cancer mutations
-
MIT scientists unveil breakthrough gene-editing screen to identify cancer mutations rapidly


Leave a Reply
You must be logged in to post a comment.